Research
What is biological noise and why is it interesting?
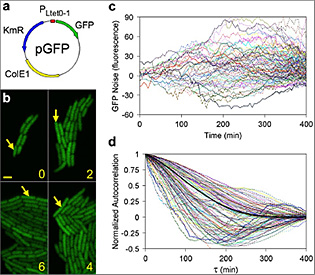
Fluctuations in gene expression (or "noise") are due to the random timing and discrete nature of biochemical interactions that occur throughout gene expression processes (i.e., transcription, translation, etc.). Gene expression noise can be used both as a probe to understand underlying gene circuit dynamics and regulation and to quantify biophysical parameters. In certain cases, noise is naturally exploited for decision making and distributing probabilities between genetic and cellular states in a cell population.
Can cellular decision-making be controlled by modulating and engineering noise? Enhanced control of decision-making at the single-cell level has importance for systems ranging from viruses and cancer to cellular reprogramming.
Fundamentals of noise and regulation of genetic circuits
To make a functional use of noise in gene expression, we must comprehensively understand the fundamentals of stochasticity and regulation of gene circuits and networks. Among several projects under this topic, the relationship between genetic architectures or structure and their function is of great interest. We have previously developed theoretical and experimental tools to understand auto-regulation and episodic transcription in E. coli (Austin, et al., 2006; Cox, et al., 2008) and infected human T-cells (Weinberger, Dar, Simpson, 2008; Dar, Razooky, et al., 2012). We also have investigated the differences in genetic architectures that couple noise and transcriptional response to perturbations in budding yeast (Dar et al., 2010). Gene circuits in E. coli, yeast, and mammalian cells are studied using experimental and computational approaches. Our lab integrates both system-wide datasets and single-cell time-lapse fluorescence microscopy.
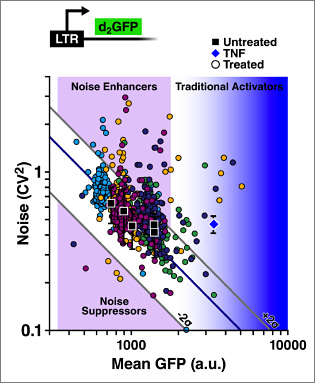
Applications of noise drug screening and engineering noise
Recently we introduced noise screening of HIV-1 gene circuitry and demonstrated that combinations of noise-modulating drug treatments enhance viral reactivation through diverse mechanisms (Dar, et al., 2014). To extend noise drug screening beyond this system, we are interested in biasing diverse cellular decisions and advancing the fundamentals and tools for noise drug screening. A new field of “noise pharmacology” will require a currently non-existent foundation in three areas:
- Sources of noise in gene expression (e.g., transcriptional, post-transcriptional, etc.) (See Megaridis et al., 2018)
- Regulatory motifs and networks (e.g., auto-regulation, noise propagation, mRNA regulation)
- Cell-fate decisions in disease phenotypes (e.g., viruses, cancer, stem cells, etc.)(See Bohn-Wippert et al., 2018)
Modulation of Stochastic Pluripotent Gene Expression
Our goal is to engineer pluripotent gene expression noise within stem cells to establish controlled methods for multi-lineage commitment. Traditional stem cell differentiation is directed towards a single specific lineage. However, our physiological tissues are composed of many cell types that work in unison to develop emergent behaviors. By modulating the amount of noise of stem cells at early stages of differentiation, it could be possible to effectively tune the simultaneous co-differentiation of multiple cell lines. One of the holy grails of stem cell research is to be able to grow fully sized, functional organs that can be transplanted into patients. Biological noise may be one of the keys that begins to unlock the possibility of reaching that goal.
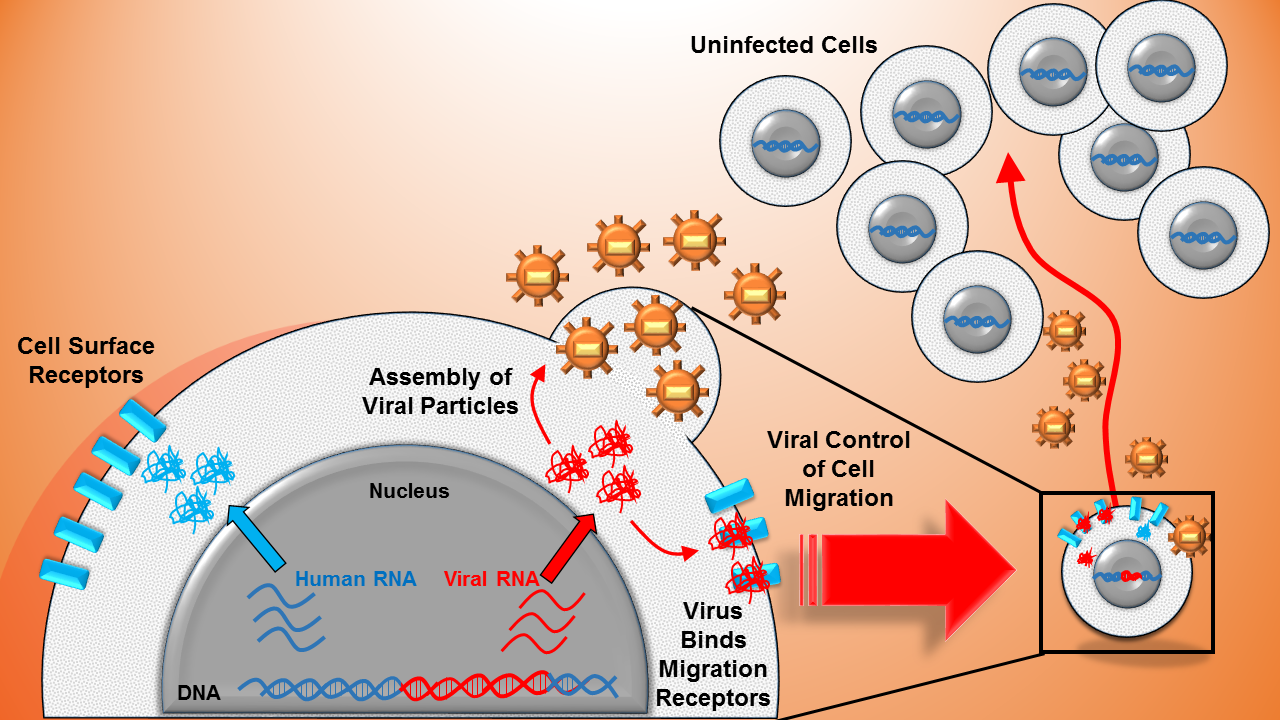
Computational and Systems Bioengineering: A gateway to reverse-engineer natural systems
In addition to studing noise, our lab is interested to integrate genome-wide and perturbation datasets with experimentation towards understanding Nature’s blueprints of cellular organization. Interests include the structure of genome-wide noise sources in E. coli (Dar et al., 2015) and studying viral-host relationships to improve therapeutic strategies (Bohn-Wippert et al., 2017).
Current openings
Graduate Students and Postdoctoral Researchers
Passionate and driven individuals are wanted for diverse projects spanning basic to translational research in mammalian systems in a highly interdisciplinary and dynamic lab environment. Projects utilize diverse modeling, experimental techniques, and collaborations. Backgrounds in Biology, Physics, Engineering, and Computer Science are welcomed.
If interested in these positions, please email a brief cover letter with research interests and CV to Dr. Dar.




